欢迎对植物发育生物学、表观遗传学研究感兴趣的本科生、研究生(硕士、博士)、博士后加入实验室!
教育经历
2001.9–2005.7 中国农业大学(CAU) 生物科学 学士
2005.9–2011.7 中国农业大学(CAU),北京生命科学研究所(NIBS) 植物学 博士
工作经历
2011.11–2017.10 英国John Innes Centre 博士后、高级研究助理
2017.11-现在 武汉大学生命科学学院 教授、博士生导师
主要研究方向
植物生殖转变的表观遗传学调控机制
1. Long non-coding RNA COOLAIR 的表达调控及其对FLC的调控机制研究
2. PRC2调控基因表达的机制
3. PRC1调控雌配子体发育的机制
4. 水稻PRC1和PRC2调控发育的机制研究
植物生殖转变研究室
生殖转变是开花植物形成种子、完成生活史的关键步骤。在此过程中植物由产生叶片的营养生长转变为开花、形成种子的生殖发育。本实验室以该过程为研究对象,探索表观遗传学调控机制,尤其是多梳蛋白复合体(PRC complex)(Fig 1),非编码RNA包括长链非编码RNA(lncRNA)和小RNA(small RNA)介导的基因表达调控在此转变过程中的重要作用。
拟南芥方面的工作
植物的生殖转变包括三个相对独立又相互联系的过程:1)植物获得开花的能力;2)花原基的分化;3)配子体的发生和发育。研究表明表观遗传学调控机制在此过程中有非常重要的调控作用。本实验室将以模式植物拟南芥(Arabidopsis)和水稻(Oryza sativa)为研究材料,综合利用遗传学、分子生物学、生物化学、细胞生物学、生物信息学等多种研究技术和手段,研究该过程中表观遗传学对重要基因表达状态的改变和组织特异性形成的调控机制,并解析这些改变对植物发育的调控作用,从而建立植物生殖转变的表观遗传学调控网络。
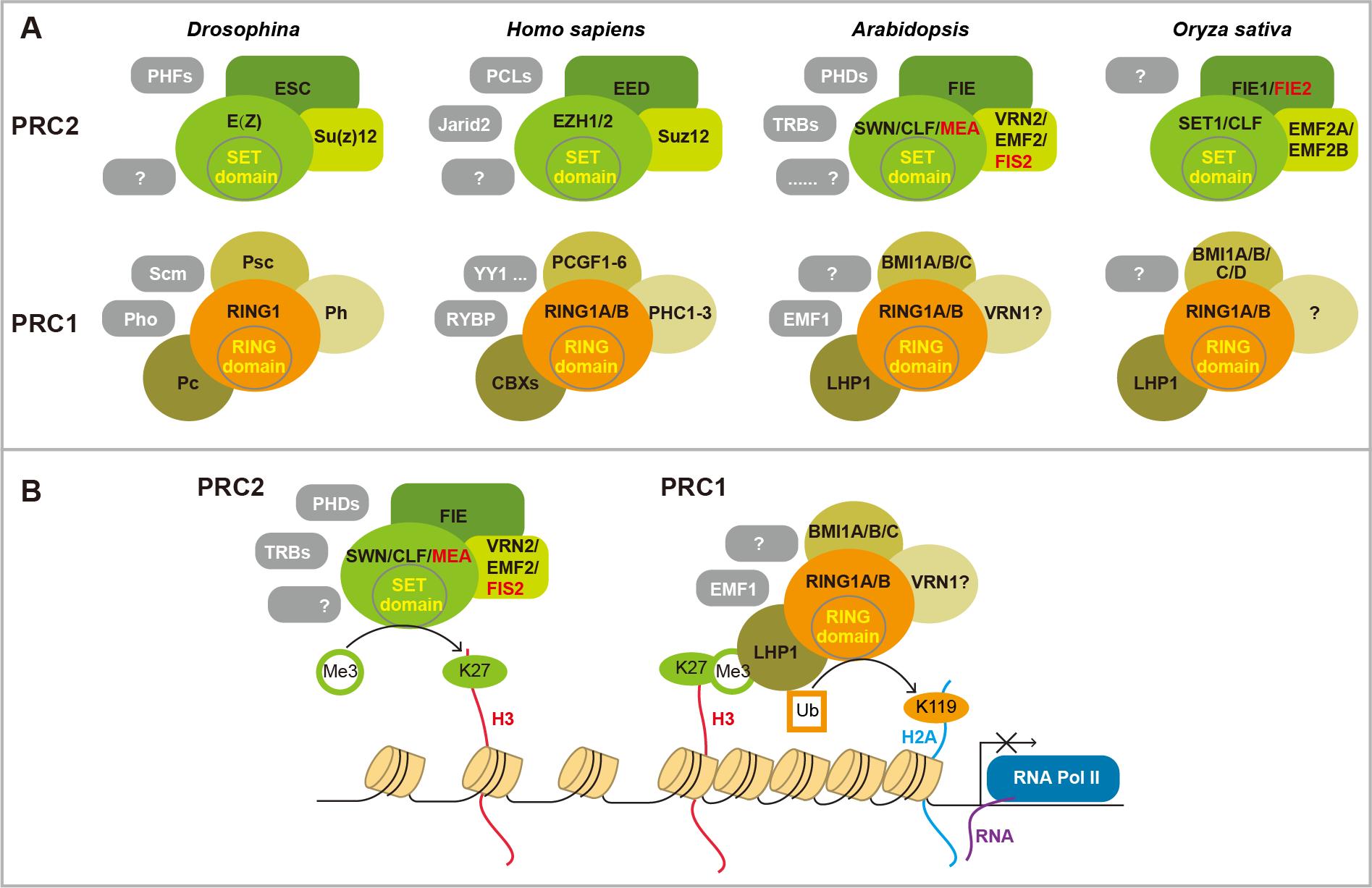
Fig 1. PRC complexes and working model.(A) There are two conserved PRC complexes PRC1 and PRC2. PRC2 is composed of three core components of a WD40 protein Esc, a Zinc-Finger VEFS protein Su(z)12, and a methyltransferase E(z) that harbors a catalytic SET domain containing E(z). A few accessory proteins (gray boxes) have been found in different organisms. These accessories specify PRC2 target genes or regulate its activity. Four core PRC1 proteins have been found in Drosophila, including an H3K27me3 reader Pc, an E3 ligase RING domain protein RING1, Psc, and Ph. Some components are lacking in plant. The accessory proteins interact with core components, and specify its activity at certain conditions. (B) Working model of PRC cascade. PRC2 methylates histone H3 lysine 27 (H3K27) to repress gene expression. Pc component in PRC1 can bind to H3K27me3. Then PRC1 catalyzes histone H2AK119 mono-ubiquitination to compact chromatin, thereby maintaining the repressed state. However, each of the complexes can function independently. Red colored components are mainly expressed in endosperm.
拟南芥开花能力的调控
拟南芥由营养生长转变为生殖生长的先决条件是植物必须具备开花的能力,在此过程中MADS-box转录因子FLOWERING LOCUS C (FLC)具有关键性的作用。FLC通过抑制下游开花素基因FLOWERING LOCUS T (FT)的表达抑制拟南芥开花。现有的研究表明很多内源因子和外部环境都可以通过调节FLC染色质的状态调节FLC的表达,进而调控开花时间(Fig 2)。
特定位点的组蛋白修饰可以调控基因的表达,组蛋白H3K4me3和H3K36me3能够促进基因的表达;而PRC2通过甲基化组蛋白H3K27,抑制基因的表达。实验室负责人前期的作表明拟南芥在越冬生长之前的常温生长过程中,组蛋白H3K4me3脱甲基化酶JMJ15和JMJ18可以降低FLC染色质上H3K4me3水平,抑制FLC的表达(Yang et al., PLoS Genet 2012; Yang et al., Plant Cell Rep 2012),促进开花;同时组蛋白H3K36甲基化酶SDG8和组蛋白H3K27脱甲基化酶ELF6通过蛋白相互作用,形成蛋白复合体,在促进H3K36me3的同时抑制H3K27me3的产生,从而促进FLC的表达,保持植物处于营养生长的状态(Yang et al., Curr Biol 2014; Yang et al., PNAS 2016)(Fig 3);在长时间低温生长也就是春化作用过程中,PRC2复合体及其附件蛋白PHD蛋白VIN3、VRN5形成PHD-PRC2复合体,使FLC染色质特定区域- nucleation region的H3K36me3转变为H3K27me3,从而抑制春化前活跃表达的FLC。PHD蛋白VIN3的表达受到春化的特异性诱导,从而保证了该过程只在春化作用下发生;春化作用之后,VIN3不再表达,VRN5-PRC2依然可以在一定时间(大约10天)内结合在FLC nucleation region,保持FLC的抑制状态,同时PRC2复合体能够从FLC nucleation region扩散到整个FLC染色质上,并通过与H3K27me3结合蛋白LHP1相互作用,形成read-write的正反馈作用,在细胞分裂过程中使H3K27me3覆盖整个FLC染色质区域,保持FLC的低表达状态,促进开花(Yang et al., Curr Biol 2014; Yang et al., 2017)。

Fig 2. Model of FLC mediated flowering time regulation.The epigenetic histone modification of each regulation pathway is indicated in black at the base of each pathway. |
Fig 3. Schematic picture of FLC epigenetic regulation in vernalization.Vernalization process is separated to three connected time period: warm growth before vernalization (Pre-cold), long term cold treatment (In the cold), and normal growth after a certain period of cold (Post-cold). |
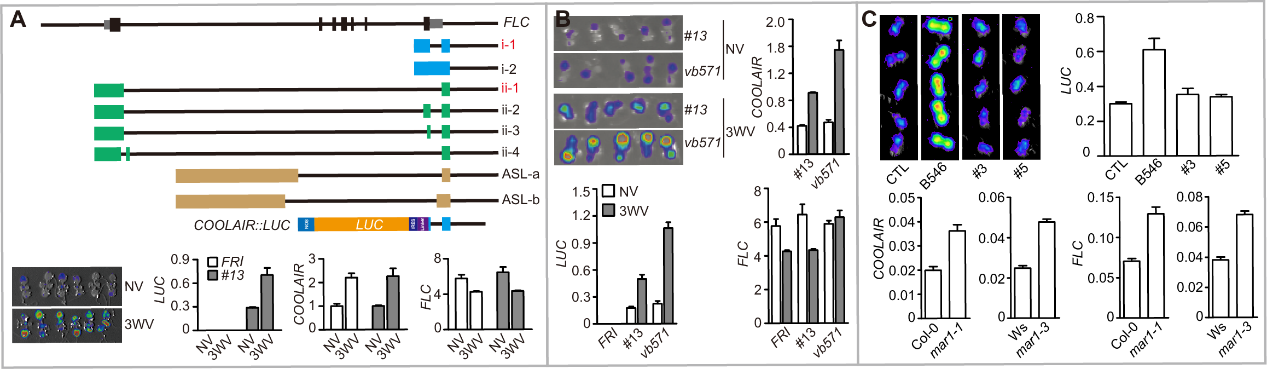
Fig 4. COOLAIR::LUC screening system. (A) FLC gene structure and isoforms of FLC antisense transcripts. The dominant COOLAIR isoforms were labeled by red characters. The schematic picture of COOLAIR::LUC construct. The mutagenized progenitor line #13 was also tested by LUC image and qRT-PCR in NV and 3WV cold treatment conditions. (B) and (C) Examples of selected mutants after EMS mutagenesis. (B) vb571 displayed a cold induced up-regulation of COOLAIR. The phenotype was determined by LUC imaging and qRT-PCR. FLC cannot be efficiently repressed in the cold. (C) A warm higher COOLAIR::LUC mutant was mapped to MAR1. The mutant phenotype is complemented by the genomic transgene. The mutant phenotype was also confirmed by another allele mar1-3 in Ws background.
研究还表明,FLC除了能够转录编码蛋白的正义(sense) RNA以外,还可以从3’末端转录出一系列的长链非编码RNA (long non-coding RNA) COOLAIR (Cold Induced Long Antisense Intragenic RNA)( Swiezewski et al., Nature 2009)和ASL (Antisense Long)(Shin et al., PLoS Genet 2014)转录本(Fig 4)。现有的研究表明,这些长链非编码RNA在常温生长和春化作用过程中对FLC sense的表达和转录有重要的调节作用(Wu et al., Plant Physiol 2020)。因此Caroline Dean实验室利用COOLAIR的启动子和部分转录区域连接报告基因LUC,构建了COOLAIR::LUC转基因植物(Fig 4)。实验表明COOLAIR::LUC和内源的COOLAIR具有相似的表达调控模式。因此COOLAIR::LUC是一个非常好的用来研究FLC antisense表达调控、及其对sense调控机制的有效工具,并且已经证明这一点(Sun et al., Science 2014)。本实验室在此基础上进行了一定的改进,并通过遗传筛选克隆了几个COOLAIR的调控因子,并检测了这些因子对FLC sense表达的影响。我们正在利用多种方法研究这些调控因子调控COOLAIR的机制,并期望回答三个主要的问题:1)这些因子是如何调控COOLAIR;2)COOLAIR如何调控FLC sense;3)对PRC2或PHD-PRC2的功能有什么影响。
花原基的分化
植物完成由营养生长向生殖生长的转变的同时,营养顶端分生组织也转变为了花序顶端分生组织。花的形成还需要花序顶端分生组织分化出花原基,在此过程中两个转录因子LEAFY (LFY)和AGMOUS (AG)发挥了关键性的作用,二者的表达状态发生了一系列的转变,形成了特定了时空表达特异性,并且这些表达状态和模式的改变直接决定了花发育的过程。
LFY和AG都是PRC2复合体的靶标基因,并调控二者的时空表达特异性(Fig 5)。我们以LFY和AG为研究对象,研究PRC2调控基因的一些关键问题:1) PRC2靶向调控基因的机制;2) PRC2从招募区域向外扩散的机制;3) PRC2建立边界的机制;4) 基因沉默和活跃表达相互转换的机制。目前在不同的物种中对这些问题都有一定的研究,尤其是PRC2对特定基因的靶向机制已经有了比较深入的研究,但对另外三个问题研究较少。
我们选取LFY和AG作为研究对象有一下几个理由:1)LFY和AG对花发育有关键性的调控作用,二者异位表达都会影响植物的正常发育;2)LFY和AG代表了两种不同的PCR2靶向调控机制;PRC2在拟南芥基因组中有>6000个靶标基因,其中只有<100基因有PRE-like (Polycomb response element like)调控序列,一些转录因子通过识别这些特定的DNA调控元件,结合到染色质上,然后通过蛋白相互作用招募PRC2复合体,完成PRC2的靶向招募(Yuan et al., Nat Genet 2016; Xiao et al., Nat Genet 2017; Zhou et al., Nat Genet 2018)。AG就是具有PRE-like DNA原件的基因。目前对于PRC2如何结合到AG染色质的特定区域已经有了比较深入的研究,但是在发育过程中和表达转化过程中,PRC2对AG的动态调控机制还研究较少。PRC2的绝大多数靶标基因都没有预测的PRE-like调控序列,LFY就是其中的典型代表,因此我们期望通过PRC2对LFY靶向机制的研究,揭示PRC2对这类基因的靶向调节机制。3)绝大多数PRC2靶标基因上H3K27me3的修饰都与周围的基因有明确的界限,LFY和AG也是如此,只有很少的研究涉及了该问题(Narendra et al., Science 2015; Yan et al., Nat Plants 2018),希望通过我们的研究,加深对该问题的认识。4)我们期望把上面的研究和具体的发育过程联系起来,揭示基因的精确调控和发育进程的关系。
我们已经针对LFY和AG设计了一些遗传筛选体系,目前正在检测这些遗传筛选体系,已经有一些比较积极的结果。
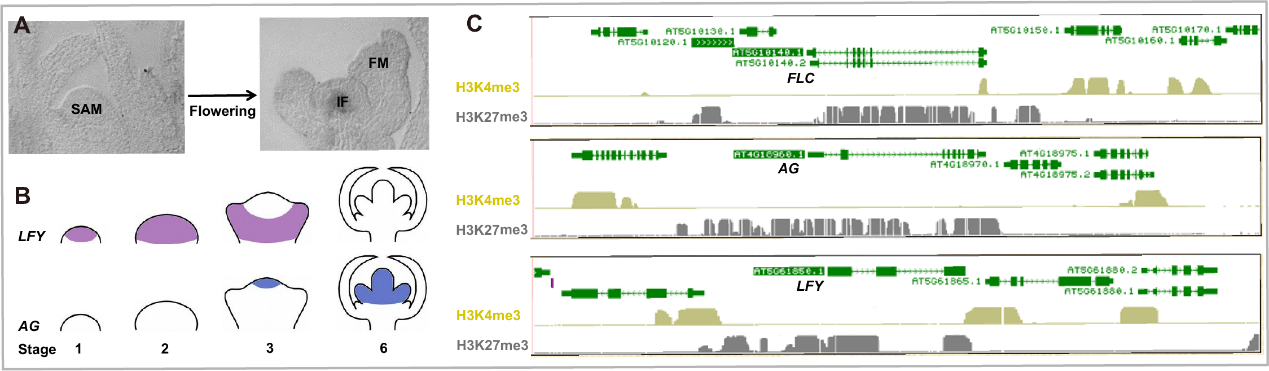
Fig 5. The expression patterns of AG and LFY in reproductive transition. (A) The meristem transition in flowering. SAM, shoot apical meristem; IF, inflorescence meristem; FM, flower meristem. (B) AG and LFY expression pattern dynamics during floral development. The colored regions indicate the expressed domains. Floral development stage was numbered at the bottom. (C) The patterns of activation histone mark H3K4me3 and repression modification H3K27me3 at FLC, AG, and LFY at seedling stage.
雌配子体的发生和发育
在花发育的过程中,一个非常重要的事件是一个或一些特定的体细胞转变为具有生殖潜力的孢子母细胞,然后进一步发育为功能孢子。这是开花植物特异的发育过程,目前对这个过程的研究还比较少。我们主要关注的是雌配子体的发生和发育。
我们现有的结果表明,多梳蛋白复合体中的PRC1能够调节该过程。在拟南芥中,PRC1有两个功能冗余的RING1的组分,RING1A和RING1B。我们得到了一个RING1A的弱突变体ring1a-2,并用此构建了ring1a-2 ring1b双突变体,该突变体有不育的表型。进一步的表型分析表明该突变体花粉的育性正常,雌配子体几乎不育。更精细的表型观察发现,该突变体中雌配子体发育的缺陷最早发生在体细胞特化为大孢子母细胞的阶段,其随后的发育也受到了极大的影响。现在我们正在通过细胞生物学和生物化学的方法研究PRC1调控该发育过程的机制。
为了进一步研究PRC1在拟南芥中的功能,我们还通过遗传筛选得到了多个ring1a-2 ring1b的抑制子,这些突变体恢复了ring1a-2 ring1b 某些和所有的发育表型。我们已经通过BSA测序分析的方法克隆了其中两个突变体,并进行了一系列的遗传分析。我们希望通过这些研究能够揭示PRC1调控植物发育的机制。
水稻方向的研究
我们也以单子叶模式植物水稻为研究材料,研究PRC1和PRC2复合体调控水稻发育的机制。目前我们构建了一系列的相关突变体和转基因材料。我们还通过酵母双杂交实验筛选了一些与PRC2组分相互作用的转录因子,并构建了相关的遗传材料。我们正在研究这些转录因子与PRC2复合体的关系,以及它们对PRC2功能的影响。
Plant Reproductive Transition Lab
Reproductive transition is the most critical step for setting seeds and finishing life cycle in flowering plant. In this switching, the plant shoot apical meristem (STM) transforms to inflorescent meristem (IF), and in turn, produces flower. We are focusing on exploring the regulation roles of Polycomb Repression Complexs (PRCs) (Fig 1) and Long Non-coding RNA in this key developmental event.
Work on Arabidopsis
The reproductive transition includes three relative independent, but tightly linked developmental processes: 1) gaining the flowering ability; 2) forming flower primordia; 3) gametophytes biogenesis and development. Our lab will use the model plants Arabidopsis and Oryza sativa to study the epigenetic regulation mechanisms underpinning these processes. We try to understand regulation mechanisms on key factors, which display tissue specificity, expression on/off transition in development. Hence, we could connect gene expression dynamics to a certain developmental process. By our effort, we will further explore and understand gene regulation networks in reproductive transition.
Flowering Transition
The MADS-box transcription repressor FLOWERING LOCUS C (FLC) is the central flowering repressor. It directly inhibits the florigen gene FLOWERING LOCUS T (FT) expression to repress flowering. Several developmental factors and environment cues have been identified to regulate FLC expression through epigenetic regulation pathways (Fig 2). Our previous work have illustrated several epigenetic regulation mechanisms on FLC in warm and vernalization conditions (Fig 3).
A set of FLC long non-coding RNA including COOLAIR and ASLs have been characterized (Fig 3), and linked to FLC sense regulation. But little is known on COOLAIR regulation. Aiming to explore mechanisms of COOLAIR regulation and FLC regulation by COOLAIR. A genetic screening named as COOLAIR::LUC screening were performed (Fig 4). The first identified mutant indicated that the DNA-RNA hybrid structure called as R-Loop is critical for COOLAIR and FLC sense regulation. We further modified the screening system and re-performed the COOLAIR::LUC screening. Several mutants have been identified and roughly characterized (Fig 4). Our further study will make significant improvement on understanding 1) COOLAIR regulation mechanism; 2) linking COOLAIR expression dynamics to FLC sense regulation; 3) whether these mechanisms link to PRC2 or PHD-PRC2 function.
Flower Primordia Development
The plant switches from vegetative growth to reproductive development. Meanwhile, the shoot apical meristem (SAM) transforms to inflorescence meristem (IF), and further differentiates to flower meristem (FM) (Fig 5). LFY and AG are key components of FM differentiation. Their expression transition dynamics and tissue specific patterns determine proper flower development.
Both LFY and AG are PRC2 targets. But they have distinct features of PRC2 targeting. There are Polycomb Response Element-Like (PRE-L) sequences in AG locus, but not in LFY. There are more than 6000 PRC2 targeted genes in Arabidopsis genome. Only very small amount of them (<150) have this cis element, the others do not. Till now, studies mainly focused on PRE based PRC2 targeting. Little is known about the recruitment mechanism without PRE. We designed several screening systems. We hope the identified mutants will help to answer several basic questions on the PRC2 target genes with or without PRE-L: 1) PRC2 recruiting mechanism; 2) spreading mechanism from the initial recruiting site; 3) generating PRC2 boundary on target genes; 4) how these regulation mechanisms control plant development. Now, we have very preliminary positive data on setting these screening systems.
Megaspore Biogenesis and Development
During flower development, the plant has to produce male and female gametophytes from somatic cells. We are more interested in megaspore biogenesis and development. By chance, we got a PRC1 mutant, which is almost sterile. Detailed phonotype analysis displayed that the pollen amount is reduced, but still fertilized. Further cell biology analysis exhibits that the megaspore mother cell (MMC) biogenesis and followed development was seriously defected. We are trying to explore the mechanism on PRC1 controlled MMC biogenesis and development.
On the other hand, we like to know more about PRC1 regulation mechanism. We performed a PRC1 suppressor screening. We are looking for mutants, which could reverse PRC1 mutant all developmental or part of developmental defects. By sequencing based BSA map cloning, two of mutants have been identified. Further characterization is taking.
Work on Rice
As a member of State Key Laboratory of Hybrid Rice, the model species Oryza sativa is also used in our study. We identified several transcription factors, which physically interact with one or several PRC2 components in rice by yeast two-hybrid. The protein interaction have been confirmed with other methods. Different genetic materials have been constructed, or are ongoing. We are looking forward to reveal PRC2 function in rice development.
实验室研究人员及研究方向

发表文章
1.Yang, H.*, Berry, S. *, Olsson, T., Hartley, M., Howard, M.#, and Dean, C.# (2017). Distinct phases of Polycomb silencing to hold epigenetic memory of cold in Arabidopsis.Science357, 1142-1145. (* Co-first authors, # co-corresponding authors).
2.Yang, H., Howard, M., and Dean, C. (2016) Physical coupling of activation and de-repression activities to maintain an active transcriptional state atFLC.PNAS113, 9369-74.
3.Yang, H., Howard, M., and Dean, C. (2014). Antagonistic roles for H3K36me3 and H3K27me3 in the cold-induced epigenetic switch at ArabidopsisFLC.Curr Biol24, 1793-1797.
4.Yang, H., Han, Z., Cao, Y., Fan, D., Li, H., Mo, H., Feng, Y., Liu, L., Wang, Z., Yue, Y., et al. (2012). A companion cell-dominant and developmentally regulated H3K4 demethylase controls flowering time inArabidopsisvia the repression ofFLCexpression.PLoS Genet8, e1002664.
5.Yang, H.*, Mo, H.*, Fan, D., Cao, Y., Cui, S., and Ma, L. (2012). Overexpression of a histone H3K4 demethylase, JMJ15, accelerates flowering time inArabidopsis. Plant Cell Rep 31, 1297-1308. (* Co-first authors).
6. Crevillen, P.,Yang, H., Cui, X., Greeff, C., Trick, M., Qiu, Q., Cao, X., and Dean, C. (2014). Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state.Nature515, 587-590.
7. Wu, Z., Ietswaart, R., Liu, F.,Yang, H., Howard, M., and Dean, C. (2016). Quantitative regulation ofFLCvia coordinated transcriptional initiation and elongation.PNAS113, 218-223.
8. Angel, A., Song, J.,Yang, H., Questa, J.I., Dean, C., and Howard, M. (2015). Vernalizing cold is registered digitally atFLC.PNAS112, 4146-4151.
9. Li, C., Xu, J., Li, J., Li, Q., andYang, H*. (2014). Involvement of Arabidopsis histone acetyltransferase HAC family genes in the ethylene signaling pathway.Plant Cell Physiol55, 426-435. (* not corresponding author).
10. Fan, D., Dai, Y., Wang, X., Wang, Z., He, H.,Yang, H., Cao, Y., Deng, X.W., and Ma, L. (2012). IBM1, a JmjC domain-containing histone demethylase, is involved in the regulation of RNA-directed DNA methylation through the epigenetic control ofRDR2andDCL3expression in Arabidopsis.Nucleic Acids Res40, 8905-8916.